The purpose of this page is to help you navigate and use PlaNet database.
1. Specifying genes for analysis.
To start using PlaNet, enter your favorite genes into the first text box available on the front page. You can use gene identifiers, probeset IDs or keywords to find your genes.
Click on search button to start the search. You can also use protein or nucleotide sequences in the box below.
2. Gene pages
After you have found and clicked on a link of your gene, you will be taken to a page dedicated to the gene. There, you will find:
1. A table containing a description of the gene. The table also contains a link to a cluster where the gene is found (discussed below) and additional information of Pfam domains and PLAZA families the gene is assigned to.
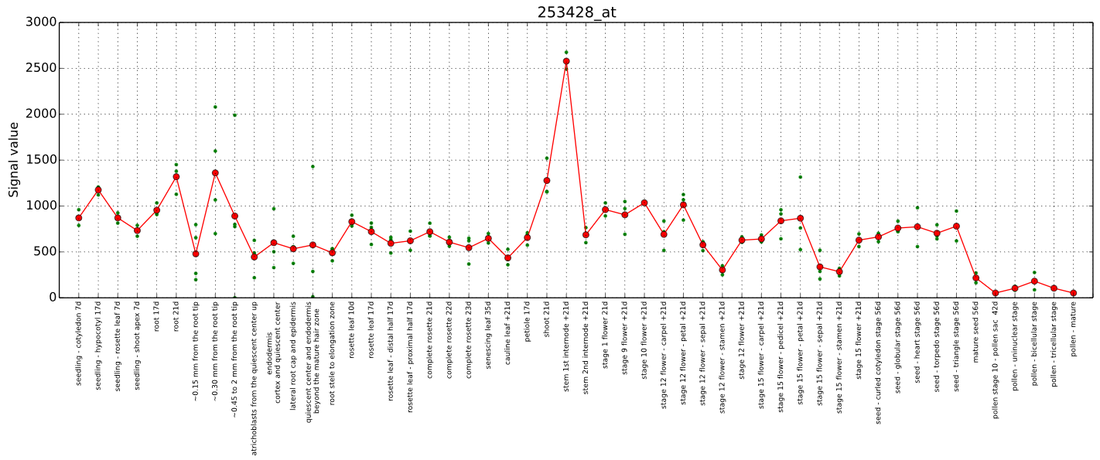
2. Expression profile of the gene. The elements on the figure are: Y-axis: expression values, x-axis: tissues/treatments, Red dots: average expression, green dots: expression from individual microarray/RNAseq experiments.
1. A table containing a description of the gene. The table also contains a link to a cluster where the gene is found (discussed below) and additional information of Pfam domains and PLAZA families the gene is assigned to.
2. Expression profile of the gene. The elements on the figure are: Y-axis: expression values, x-axis: tissues/treatments, Red dots: average expression, green dots: expression from individual microarray/RNAseq experiments.
3. Co-expression network of the gene. This network contains the query gene (large node) and all genes two steps away from the query gene. Node shapes and colors represent label co-occurrences, which are used to group the genes by Pfam domains and PLAZA families. You can right-click on the network to access a menu, where you can toggle second neighborhood, re-calculate layout and export the network as pdf. A new feature of the tool is to include ELA network (explained below in FamNet section) to highlight genes supported by the ELA.
4. Description of label co-occurrences, which are used to group genes according to the Pfam domains and PLAZA families. Each colored shape displays which labels (domains and families) are assigned to the label co-occurrence.
5. A table containing genes found in the co-expression network. The table also shows annotations and labels associated with the genes. You need to click on "Click me to toggle table containing genes in the co-expression network" to expand the table.
6. Gene Ontology enrichment analysis of the genes found in the co-expression network, where only significantly enriched terms that are present at least twice in the network are shown.
7. Gene module network that shows other networks that are significantly similar to the co-expression network of the query gene. Nodes represent gene modules found in the different species. Hover the mouse pointer over an edge to see how many label co-occurrences two modules have in common and the identity of the common labels. Right-click on the network to recalculate the layout, adjust the density, show/hide the species, show/hide conserved modules and export the pdf file of the network. Note that some genes have unique co-expression networks and will not be similar to other networks.
6. Gene Ontology enrichment analysis of the genes found in the co-expression network, where only significantly enriched terms that are present at least twice in the network are shown.
7. Gene module network that shows other networks that are significantly similar to the co-expression network of the query gene. Nodes represent gene modules found in the different species. Hover the mouse pointer over an edge to see how many label co-occurrences two modules have in common and the identity of the common labels. Right-click on the network to recalculate the layout, adjust the density, show/hide the species, show/hide conserved modules and export the pdf file of the network. Note that some genes have unique co-expression networks and will not be similar to other networks.
Edge style indicates how similar the modules are to one another, expressed as number of shared label co-occurrences (LC). Edge colors depicts the type of similarity.
8. A table containing the gene modules. If you want to see the similarities and contents of the gene modules, select the modules you want to analyze and click on "Compare".
You can specify additional parameters to control the size of the modules.
You can specify additional parameters to control the size of the modules.
- Ticking the first selection will look at first neighborhoods only; since the co-expression networks for each gene are based on second neighborhoods, this option will decrease the sizes of modules.
- Ticking the second selection will show all genes present in the selected modules; since the standard analysis shows only genes that are similar in the selected modules, this will increase the sizes of selected modules.
- Finally, the third selection will only show genes that are supported by ELA (explained below). Since the standard analysis shows all genes that are similar between selected modules, this analysis will decrease the size of selected modules. On the other hand, the genes that are supported by ELA are more likely to be biologically relevant.
3. Cluster pages
The clusters are obtained by running the genome-wide co-expression networks through HCCA algorithm. The algorithm assigns genes to clusters, which should represent densely connected regions in the co-expression network. Cluster pages are very similar to gene pages, and contain (i) interactive co-expression networks of genes found in the cluster, (ii) a gene table with annotations of the genes and (iii) Gene Ontology enrichment analysis of the genes.
4. NetworkComparer
This tool is being phased-out and replaced by FamNet.
5. FamNet
FamNet database is a successor of NetworkComparer.
Goals of the database are:
FamNet is integrated into the PlaNet database, and it's features can be found on:
Starting the analysis
To start using FamNet, navigate to FamNet page and enter gene identifier, Pfam/PLAZA identifier or a keyword (e.g. photosystem, kinase) into the text box and click "search".
To exemplify FamNet, let's use a subunit of photosystem I, PsaD family, as example. After entering "psad" into the search box, all genes that contain "psad" in the gene ID, annotation or label annotation are listed. The table also contains additional information, such as gene ID, gene description, species of the gene, and found labels.
Goals of the database are:
- to show significant, conserved associations between gene labels (gene families, and protein domains). The associations are shown as ELA networks, which are used to...
- ...find gene modules (regions of co-expression networks) that are conserved (found in multiple species) and/or duplicated (found multiple times in one species).
FamNet is integrated into the PlaNet database, and it's features can be found on:
- gene pages (see above for description of gene module networks found on these pages).
- label-specific pages.
Starting the analysis
To start using FamNet, navigate to FamNet page and enter gene identifier, Pfam/PLAZA identifier or a keyword (e.g. photosystem, kinase) into the text box and click "search".
To exemplify FamNet, let's use a subunit of photosystem I, PsaD family, as example. After entering "psad" into the search box, all genes that contain "psad" in the gene ID, annotation or label annotation are listed. The table also contains additional information, such as gene ID, gene description, species of the gene, and found labels.
By clicking on PsaD link found in the "Labels present" column, you will be redirected to a page dedicated to the label.
FamNet page for PsaD
The page contains several features.
1. A table containing description of the label.
2. A table containing genes that contain the label (PsaD). The table contains additional information, such as annotation, species and identity of other labels found in the genes.
3. ELA (Ensemble Label Association) network of the label. This network is a graphical representation of associations between different labels. The network can therefore inform you which other gene families and domains your gene family/domain is significantly associated with in multiple species. By right clicking on the network, you can recalculate the layout, export the network as pdf, or control the density of the network by selecting in how many species a given association needs to be found in. Below you can see an example of the PsaD ELA, where the influence of the different cut-offs is demonstrated. Not surprisingly, PsaD is associated with other members of the photosystem, which becomes clear as the species cut-off is increased.
FamNet page for PsaD
The page contains several features.
1. A table containing description of the label.
2. A table containing genes that contain the label (PsaD). The table contains additional information, such as annotation, species and identity of other labels found in the genes.
3. ELA (Ensemble Label Association) network of the label. This network is a graphical representation of associations between different labels. The network can therefore inform you which other gene families and domains your gene family/domain is significantly associated with in multiple species. By right clicking on the network, you can recalculate the layout, export the network as pdf, or control the density of the network by selecting in how many species a given association needs to be found in. Below you can see an example of the PsaD ELA, where the influence of the different cut-offs is demonstrated. Not surprisingly, PsaD is associated with other members of the photosystem, which becomes clear as the species cut-off is increased.
4. Gene module network that shows network regions containing the PsaD label, that are significantly similar to one another and supported by the ELA. Nodes represent gene modules found in the different species. Hover the mouse pointer over an edge to see how many label co-occurrences two modules have in common and the identity of the common labels. Right-click on the network to recalculate the layout, adjust the density, show/hide the species, show/hide conserved modules and export the pdf file of the network.
5. A table containing the gene modules. If you want to see the similarities and contents of the gene modules, select the modules you want to analyze and click on "Compare".
Selecting modules for analysis
You can specify additional parameters to control the size of the modules. Similarly to the gene pages,
Selecting modules for analysis
You can specify additional parameters to control the size of the modules. Similarly to the gene pages,
- ticking the first selection will look at first neighborhoods only; since the co-expression networks for each gene are based on second neighborhoods, this option will decrease the sizes of modules.
- Ticking the second selection will show all genes present in the selected modules; since the standard analysis shows only genes that are similar in the selected modules, this will increase the sizes of selected modules.
- Finally, the third selection will only show genes that are suppored by ELA (explained below). Since the standard analysis shows all genes that are similar between selected modules, this analysis will decrease the size of selected modules. On the other hand, the genes that are supported by ELA are more likely to be biologically relevant.
It seems that there are two PsaD modules (module 3 and 6) that are duplicated and significantly similar within Arabidopsis. To see what genes are present in the modules, let's select the two duplicated modules, and include module 1 (from Poplar) as well. To only show genes that are supported by ELA (this will decrease the number of genes shown in the next figure), the third check-box was ticked.
6. After clicking "Compare", the analysis takes some time to complete. The calculation time can range from seconds to a minute, and depends on the number of species that were selected.
The first item shown on the page are the three modules. Each box represents a module, and nodes within each module indicate genes.
6. After clicking "Compare", the analysis takes some time to complete. The calculation time can range from seconds to a minute, and depends on the number of species that were selected.
The first item shown on the page are the three modules. Each box represents a module, and nodes within each module indicate genes.
Some nodes between module 3 and 6 are connected by red dashed edges. This indicates that the modules are overlapping to some extent, as the connected genes are found in both modules. Still, the analysis found the two modules to be significantly similar, even when overlapping nodes are removed. To remove the overlapping genes, right-click on the network and select "Toggle overlapping genes". This option removes the connected nodes in module 3 and 6.
It seems that module 3 is the larger than module 6, and that module 3 it is highly similar to the poplar module 1. A conclusion from this analysis indicates that module 3 might be the main module involved in photosynthesis; the function of module 6 is unknown.
7. The next item on the page contains a legend figure with annotation of the label co-occurrences. The figure can be downloaded as pdf.
It seems that module 3 is the larger than module 6, and that module 3 it is highly similar to the poplar module 1. A conclusion from this analysis indicates that module 3 might be the main module involved in photosynthesis; the function of module 6 is unknown.
7. The next item on the page contains a legend figure with annotation of the label co-occurrences. The figure can be downloaded as pdf.
8. The purpose of the next figure is to simplify the first network. Here nodes represent label co-occurrences and node text indicates which genes are assigned to the labels. The red dashed edges indicate which label co-occurrences are overlapping, and the number of the edges indicates how many genes are actually overlapping.
9. The next figure indicates the expression profile of the module centers, i.e. the genes that you have selected on the previous page. Not surprisingly, the expression profiles indicate that the genes are only expressed in green tissues. Also, modules 3 and 6 have very similar expression profile, with some differences.
10. The table below lists the gene IDs, annotation and labels of genes found in the three modules.
11. Finally, the last table shows GOslim term enrichment analysis, which indicates that all three modules are involved in photosynthesis.
11. Finally, the last table shows GOslim term enrichment analysis, which indicates that all three modules are involved in photosynthesis.
FAQ (frequently asked questions)
Q: Nothing happens, when I click "EXPORT: as pdf". What can I do?
A: You are probably using Explorer as a browser. Please any other browser, such as chrome or firefox, or download networks in cytoscape format.
Q: The interactive networks are not visible. What's up with that?
A: The networks require Adobe Flash to operate. Make sure that it is installed and up to date.
Q: Nothing happens, when I click "EXPORT: as pdf". What can I do?
A: You are probably using Explorer as a browser. Please any other browser, such as chrome or firefox, or download networks in cytoscape format.
Q: The interactive networks are not visible. What's up with that?
A: The networks require Adobe Flash to operate. Make sure that it is installed and up to date.














