This tutorial explains how PlaNet was used to arrive at the PIN module example presented in our "Beyond genomics: studying evolution with gene networks". We assume that you have familiarized yourself with how to use PlaNet. If this is not the case, please visit the Help page. If you are still stuck, please write the corresponding author for further help.
We started the analysis by searching for pp1s18_186v6.1 (PIN from Physcomitrella), on the front page of PlaNet.
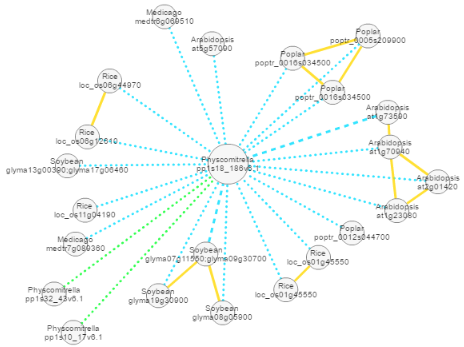
After navigating to the gene page of pp1s18_186v6.1, we scrolled to the bottom of the page, which contains a module network. The network shows if there are co-expression networks in other species (i.e. conserved modules) that are similar to the co-expression network of pp1s18_186v6.1. The analysis revealed that modules similar to the PIN module from Physcomitrella can be found in other species (conserved modules connected by blue edges to pp1s18_186v6.1).
Interestingly, we observed that Physcomitrella contains two additional PIN-like modules, indicating that these modules might be duplicated (duplicated modules connected by green edges).
Interestingly, we observed that Physcomitrella contains two additional PIN-like modules, indicating that these modules might be duplicated (duplicated modules connected by green edges).
To confirm that these duplicated modules are real, we have selected the three Physcomitrella PIN modules in the table found below the network, and clicked on "Compare".
On the bottom of the next page, the expression profiles of the selected modules are shown. The expression profiles indicate that while there are minor differences in the expression of the three modules, they are expressed in similar tissue types. Based on this information, we concluded that the two additional Physcomitrella modules are not truly duplicated.
By going back to the gene page of pp1s18_186v6.1, we observed that there are in total five PIN modules present in Arabidopsis thaliana. Since some of the modules can be overlapping (connected by an orange edge, i.e. the genes in these modules are also co-expressed to a degree), we chose to compare the two non-overlapping modules (At1g73590 and At2g01420) and the separate module (At5g57090).
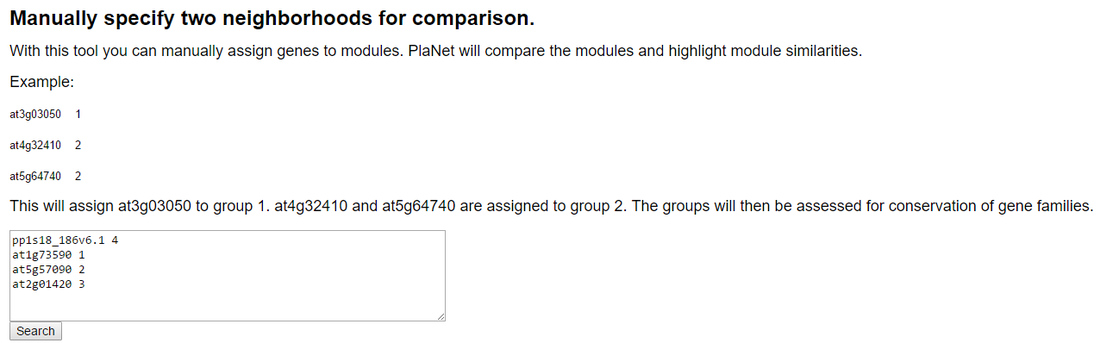
The three Arabidopsis modules, together with pp1s18_186v6.1, were entered into the module comparison tool found in the Network-comparer page (found on the top of the PlaNet pages). To specify that these four modules represent separate entities, we gave each module a unique identifier, and clicked on Search.
The next page shows which genes are present in the selected modules. The boxes represent gene modules, while nodes found in the modules represent genes. The colored shapes of the nodes indicate label co-occurrences (i.e. genes that belong to same gene families and contain same Pfam domains). The colored borders of the nodes indicate which phylostratum a given gene is assigned to, while colored edges between the modules indicate the duplication/speciation events between modules (solid edges/dashed edges, respectively). The color of these edges also correspond to the phylostratum of the duplication/speciation events. Note that the node color and layout algorithm generates the colors and layout on the fly, and the nodes need to be manually arranged to produce a network found on Figure 2.
The table below the modules indicate which phylostrata are enriched within each module. For all four modules, the Land Plant phylostratum is significantly enriched (P<0.05). Significantly depleted phylostrata are indicated by P<-0.05.
At the bottom of the page, the expression profiles of the four modules are shown. This information was used to generate the expression heat maps found on Figure 2A.
To conclude, the gene module network, phylostratigraphic analysis and expression profile information can be used to detect the gene modules, and propose their evolution.